news model
license
migration notice
This case study shows how to assess the functionality of a variant using MitImpact. It will pass through all sections of a typical result page and will explain any possible conclusion.
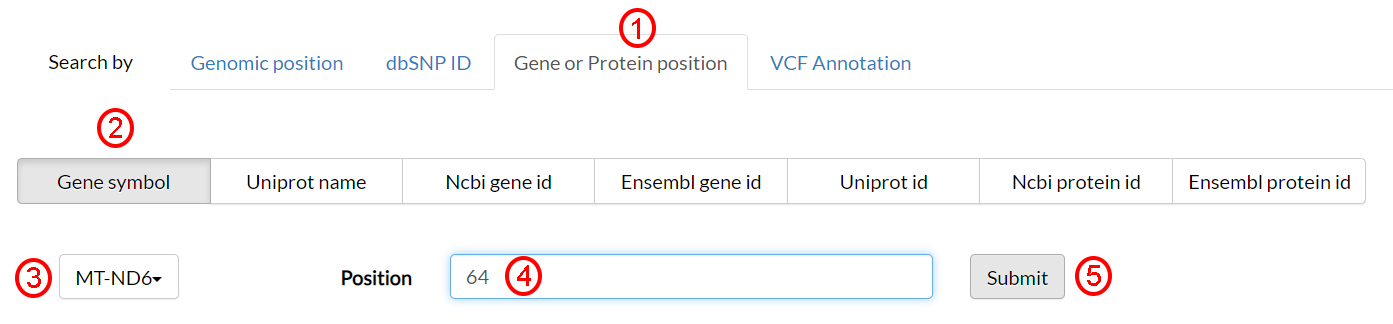
Starting from a protein of interest and an amino acid position, any associated information can be sought by selecting Gene or protein position (1). Then select Gene symbol (2) and MT_ND6 (3) from the option menu. In the Position text field (4), type 64, which is the amino acid position, and finally click on Submit (5).
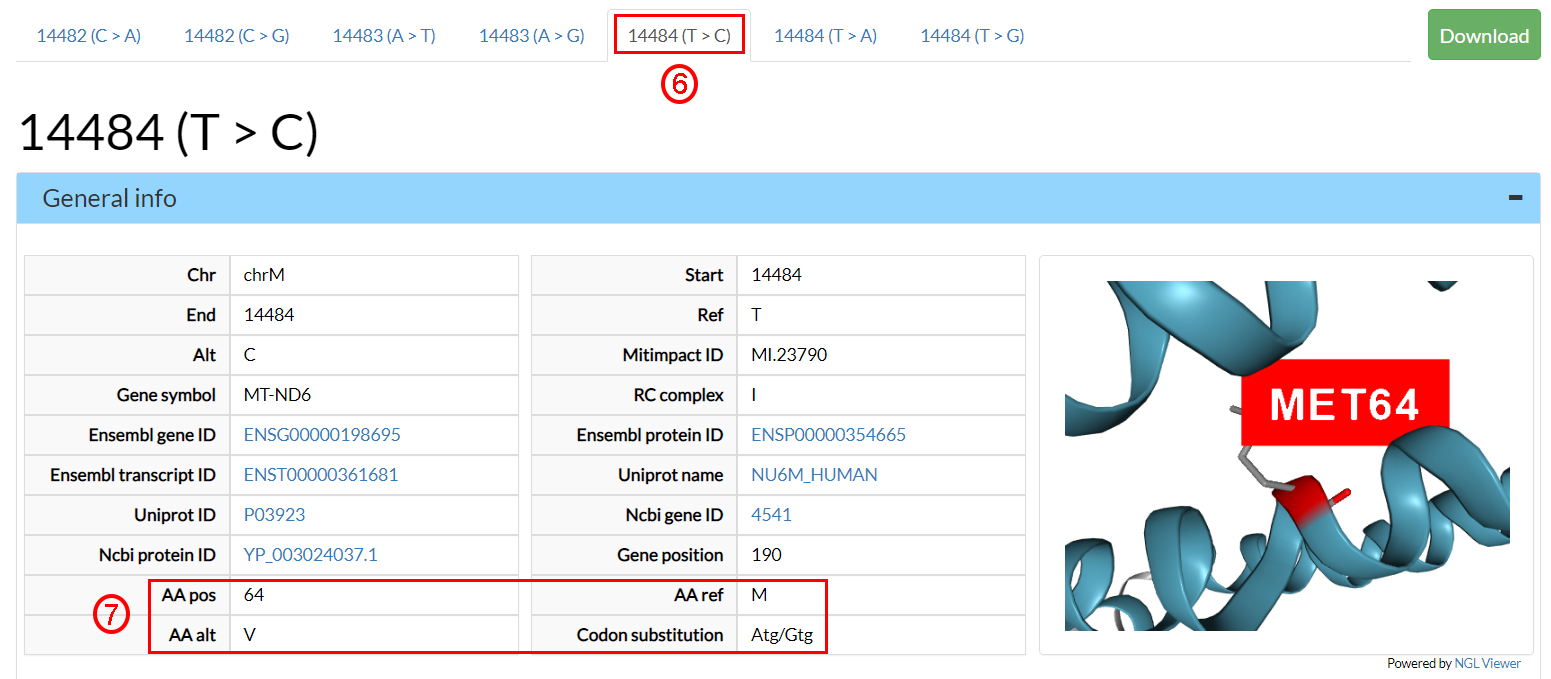
In the next page, results are split into different sections and all nucleotide variants possibly occurring
in position 64 are displayed in consecutive sheets.
Select the sheet corresponding to 14484 (T>C) (6).
In the General info section you can find several
annotations, including the alternate amino acid (7).
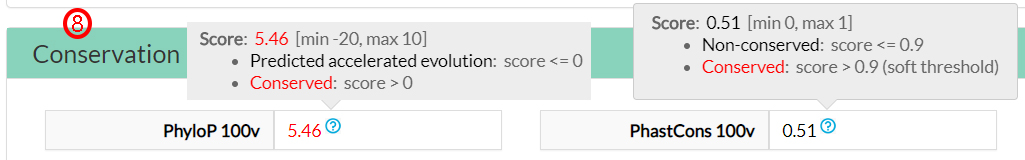
The Conservation scores indicate contrasting results depending on the score considered to measure the evolutionary conservation of this amino acid (8).
Five predictors on 16 agree that this variant amino acid (64 Valine) could be damaging (9).
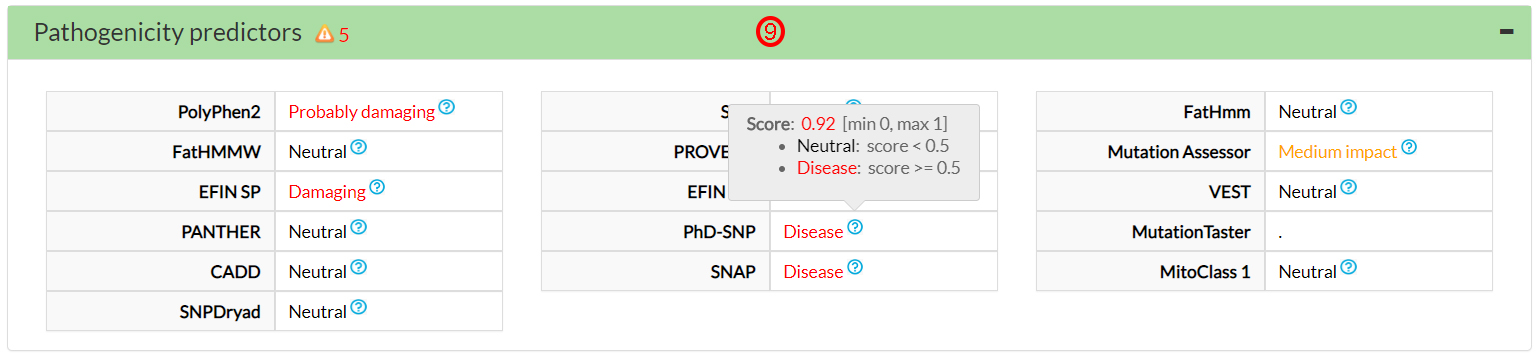
This is confirmed by the Pathogenicity meta-predictors (10) and by 2 cancer-specific predictors (11).
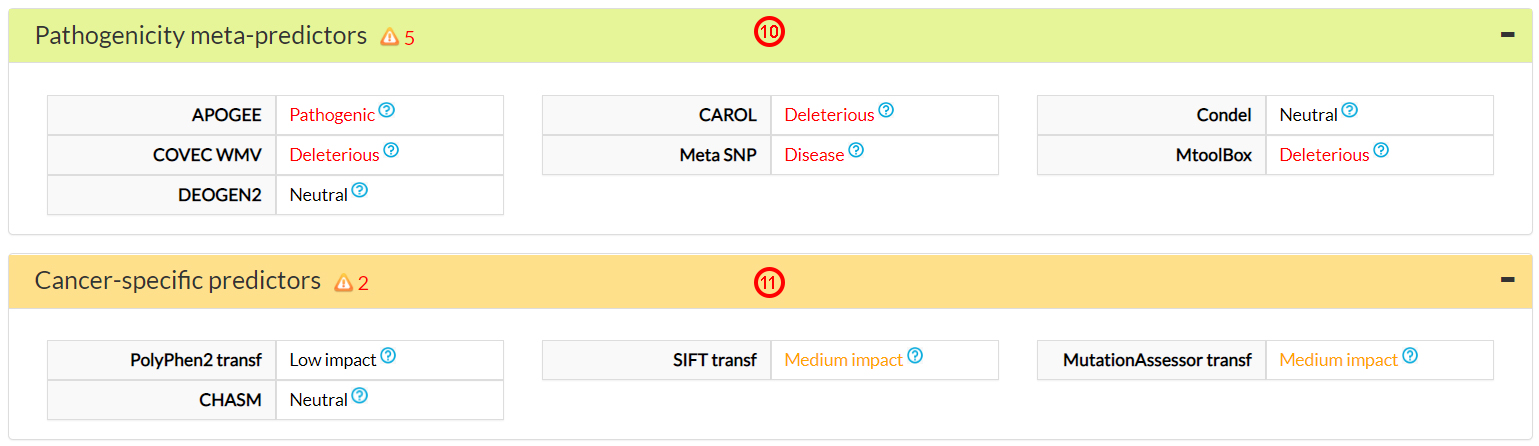
In the Databases of Phenotypes, you can find information about the association between the M64V variant and the clinical phenotypes retrieved from Clinvar and MITOMAP databases. This variant is associated with Leber’s optic atrophy and Leigh syndromes (12).
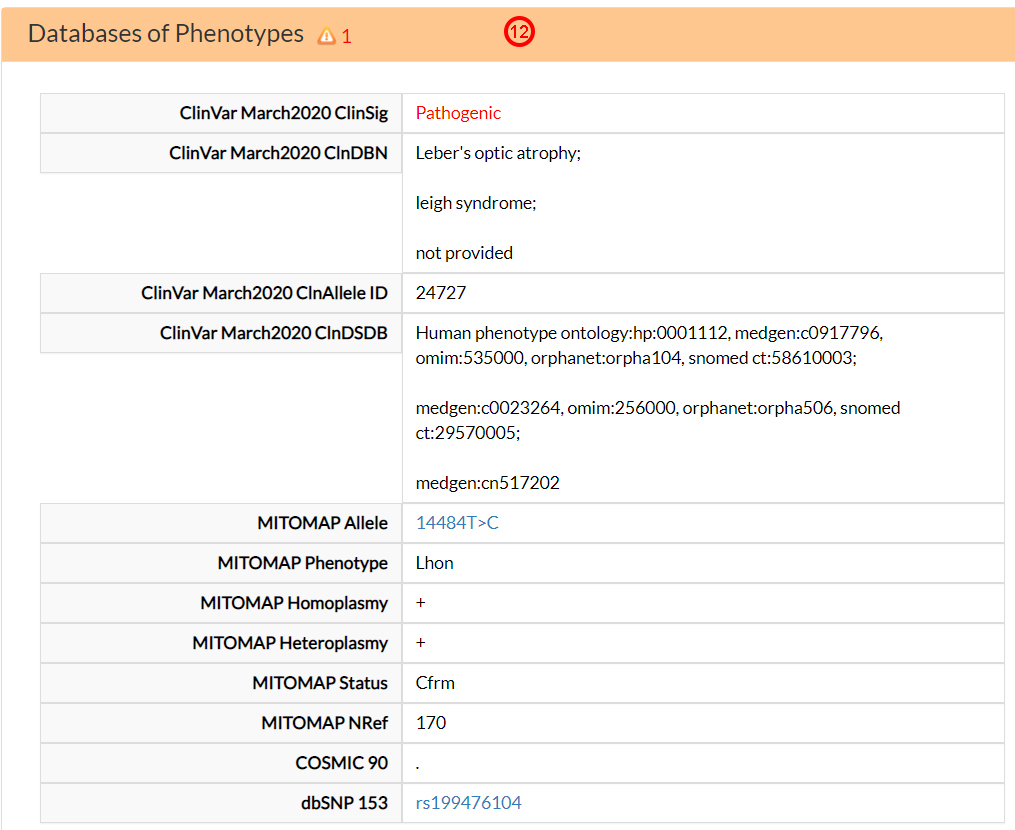
In the Residue interaction section, you can find
information on epistasis related mechanisms, specifically based on co-variation (EVmutation,
Site A-B InterP and Site A-B IntraP) and energetic parameters (DDG intra, DDG Intra interface
and DDG inter).
From the EVmutation option menu, you can select one of six predicted intra-protein (ND6) co-varying sites
(13). From the Site A-B InterP option
menu, you can find two possible covariant amino acids in two different mitochondrial proteins (ND1 and ND3)
(14). Similar to EVmutation, from the
Site A-B IntraP, you can obtain a pair of sites (positions 64 and 15) that are estimated to
co-vary (15).
It is important to remark that co-variation scores (i.e., EV score, mfDCA, and
corrected MI) are not meant to prioritize one pair over another, since all reported pairs of co-varying
variants were selected from the top scoring pairs obtained as explained in the
Description page.
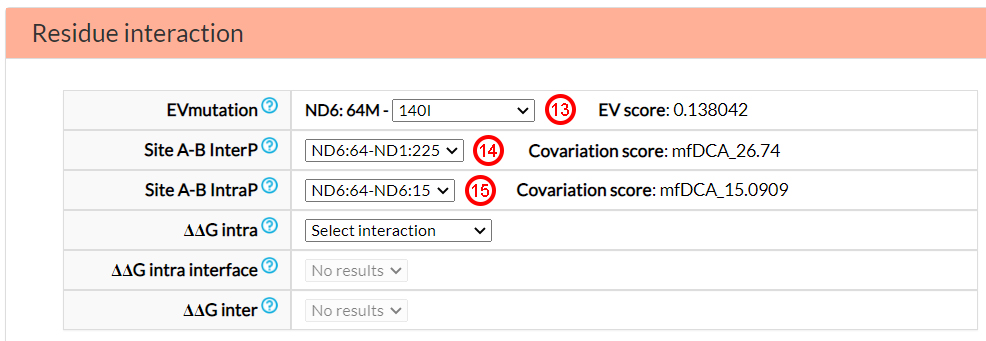
The last three menu options in the previous figure report energetic variations (DDG) of variant amino acids
at co-varying sites located inter or intra-protein. In this particular case, the variant amino acid 64V interacts
with five different variant amino acids in position 15 of the ND6 protein. Selecting. for example, the 15R
variant (16), you can obtain information on the
energetic variation caused by each single variant (DDG 1st and DDG 2nd) and of both variants
(DDG both, i.e., M64V and G15R), if occurring together (17).
A DDG both value, greater than that of the single
mutants (1.89 and 3.49), can be associated to a possibly harmful interaction between the two
variants. On the contrary, a DDG value of the double mutant close to zero can associate to an energetic
compensatory interaction. The more extreme, positively or negatively, a DDG value, the more energetically
destabilizing or stabilizing a variant can be considered.
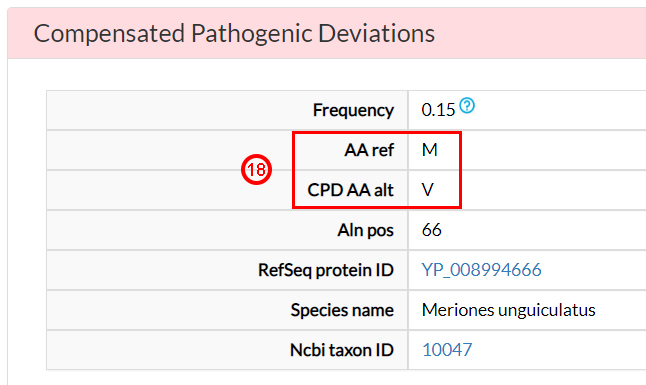
In the Compensated Pathogenic Deviations section,
the amino acid substitutions that are reported as pathogenic in the human population, but occurring as wild-type
residues in non-human ortholog proteins, are reported. These are tagged as potentially compensating
variants.
For the M64V variant, there is a potential compensating
variant (18) in at least
one species, the Meriones unguiculatus.
The deleterious effect of the variant under examination (M64V) depends on both the specific amino acid change
and the position of this residue in the ND6. Inter-protein and intra-protein co-variation analysis showed
how this site co-varies with ND1, ND3, and ND6 in position 225, 24, and 15, respectively. This result
underlies M64 as an important site of the ND6 protein since it may play a role inside a more complex
organization of the OXPHOS system.
The energetic and structural analysis of the co-varying sites demonstrate how this specific substitution
significantly destabilizes the protein and its interactions, even if the amino acid is not much evolutionary
conserved. Finally, the occurrence of a compensating pathogenic variant for M64V suggests that interacting
amino acids could modulate the deleterious effect of this amino acid variant.