news model
license
migration notice
This case study shows how to assess the functionality of a variant using MitImpact. From the Genomic position (1), search the position 14582 (2) and then click on Submit (3).
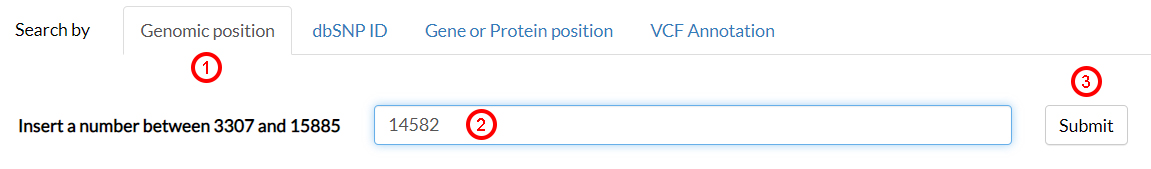
Different sections are displayed grouped in tabs on the result page, one for each possible nucleotide variant in the triplet containing the searched amino acid. Select the sheet corresponding to 14582 (A>G) (4). You can find several annotations in the General info section, including the alternate amino acid (5).
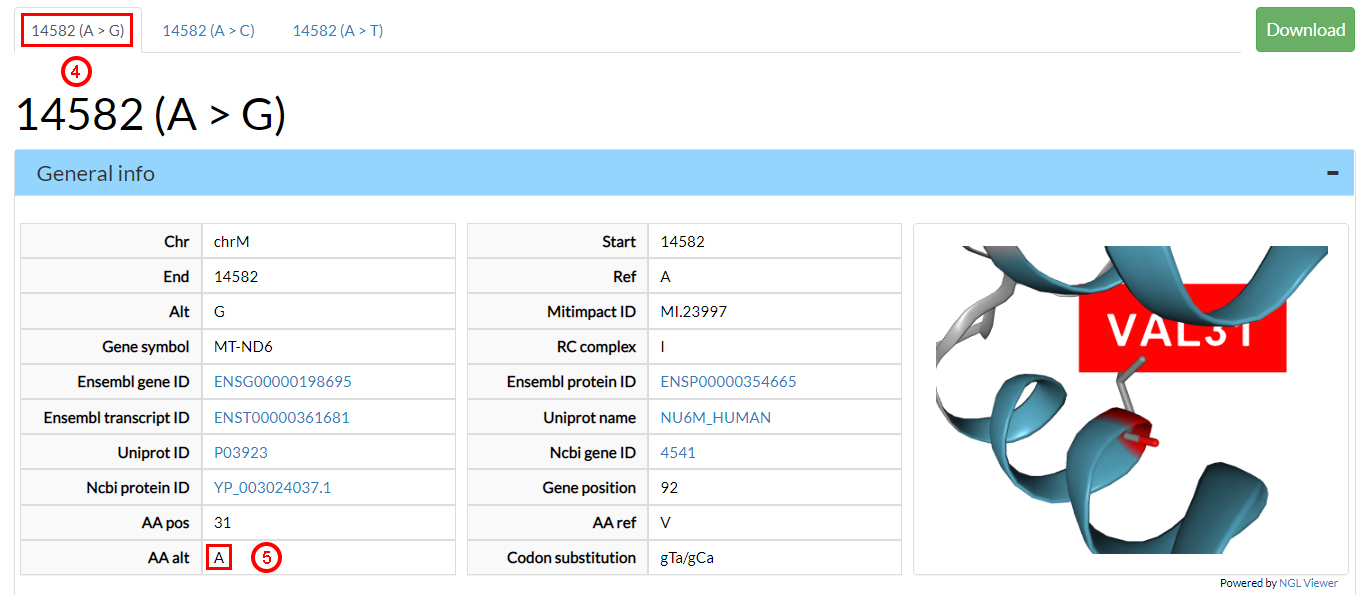
The Conservation scores indicate that this amino acid is not evolutionary conserved (6).
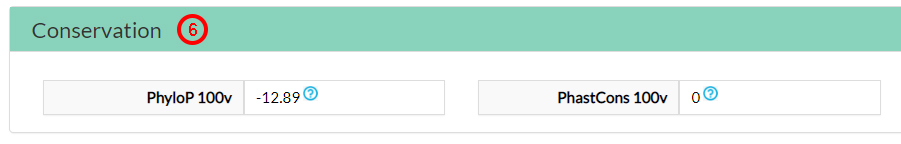
The Pathogenicity predictors suggest that this variant amino acid (31 alanine) is neutral (7).
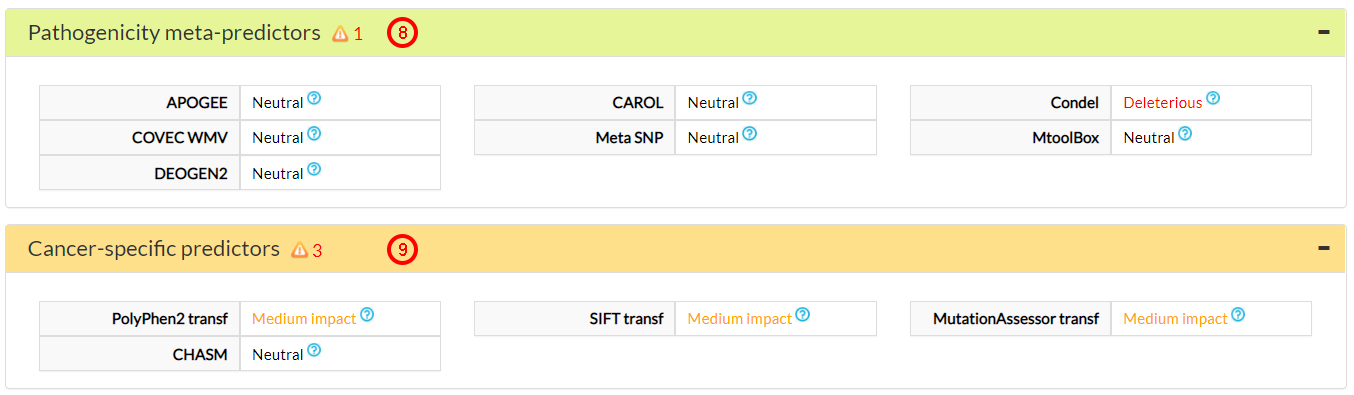
Only one Pathogenicity meta-predictor (8) and 3 cancer-specific predictors (9) suggest a potentially deleterious effect for this variant.
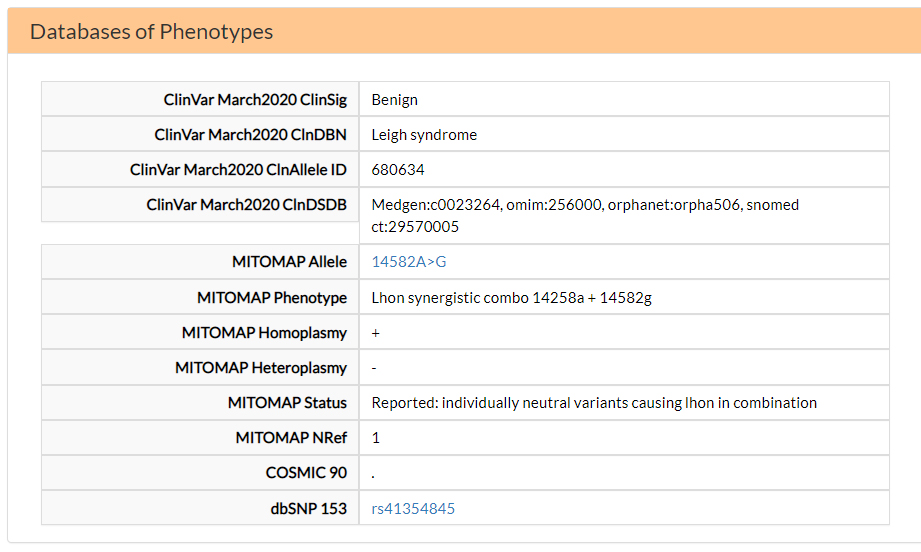
In the Databases of Phenotypes section, you can find information based on Clinvar and MITOMAP annotation databases. The V31A variant is annotated as benign in ClinVar and associated with LHON in combination with a second variant in position 14258 (Lhon synergistic combo 14258a + 14582g) in MITOMAP.
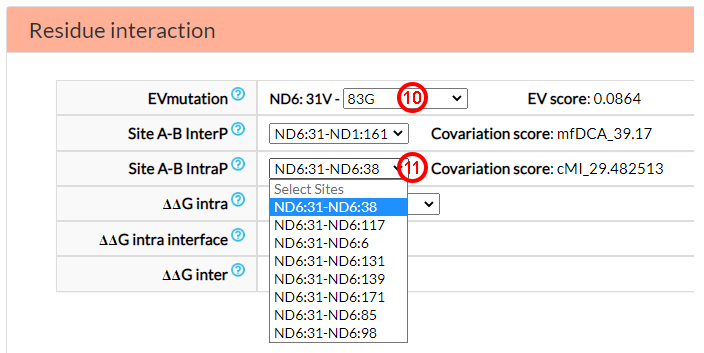
In the Residue interaction section, you can find information on epistasis related mechanisms,
specifically on co-variation (EVmutation and Site A-B, InterP and Site A-B IntraP)
and energy parameters (∆∆G intra, ∆∆G Intra interface and ∆∆G inter).
From the EVmutation option menu, you can select one of six predicted co-variant sites located in
the same protein (ND6) (10). From the Site A-B InterP
option menu, you can find 22 possible co-variant amino acids in 5 mitochondrial proteins
(ND1, ND3, ND4, ND4L, and ND5). Site A-B IntraP reports
8 co-variant residues (11).
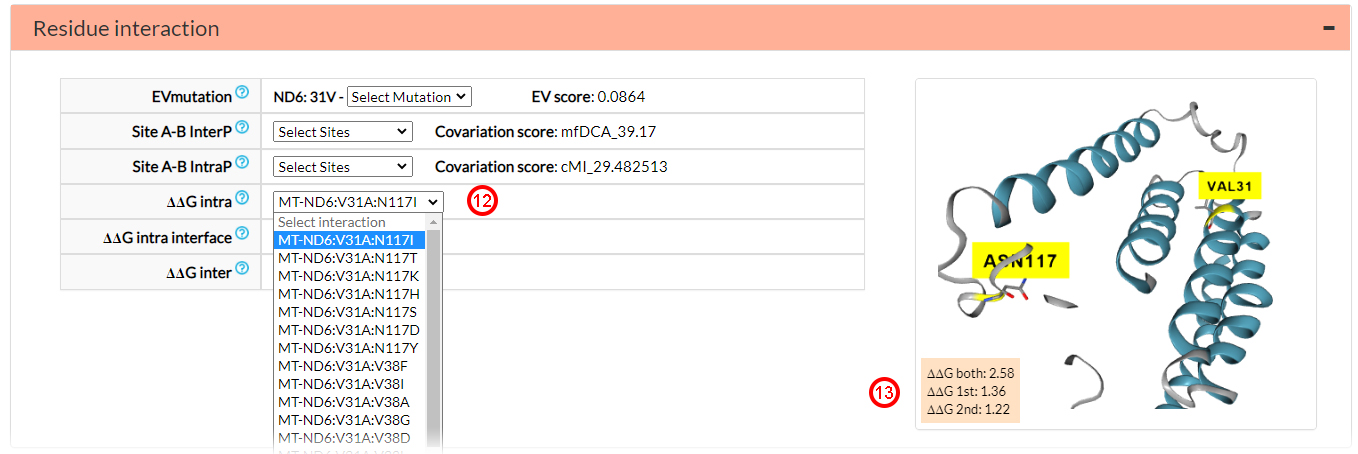
In the ∆∆G section, you will find data on co-variant sites based on energy analyses. In ∆∆G Intra you can select one among several different variant amino acids in different positions for which there are information on energy values (12). For example, If you select the N117I variant, you will retrieve the following values: ∆∆G both: 2.58, ∆∆G 1st: 1.36 and ∆∆G 2nd: 1.22 (13). There are no energy analysis results corresponding to the intra-protein interactions of residues located at the interface (∆∆G intra interface). There are no energy analysis results corresponding to interactions of residues located in different proteins (∆∆G inter)
In the Compensated Pathogenic Deviations section, you can find information on CPDs, that are
occurrences of amino acid substitutions that are reported as pathogenic in the human population,
but that occur as wild-type residues in non-human ortholog proteins.
For the V31A variant, there is not any potential compensating variant (23).
The nucleotide variant A>G in position 14582 causes the amino acid change of a residue (Valine > Alanine) in position 31 of the ND6 protein. This is not evolutionary conserved, with the pathogenicity predictors suggesting mostly a neutral effect. The co-variation sites analyses suggest the presence of several sites that may co-vary, localizing both in the same and in other mitochondrial proteins. Specifically, among the intra-protein co-varying sites, there is the Proline amino acid in position 139 that may be evolutionary linked with the residue 31. This is supported by a recent study reporting that the combination of the variants A>G in position 14582 (V31A) and G>A in position 14258 (P139L) in ND6, individually known as neutral variants, would act as synergistic variants leading to reduced OXPHOS efficiency and causing LHON.